Dr. Kenji Kozai (2020, 2021)
Consider the abstract graph K6, consisting of six vertices with edges connecting all possible pairs of vertices. We can realize the graph in space by considering the vertices as distinct points in R3 and connecting pairs of points by arcs so that no two arcs intersect. Such a realization is called a spatial embedding of the graph. A remarkable fact about K6 is that every spatial embedding contains a pair of disjoint cycles (loops) which are linked. The proof of this fact requires little more than drawing a diagram for the graph, assigning numbers +1 or -1 to crossings in the diagram to compute linking numbers, and adding up numbers modulo 2.
The red and blue cycles in this embedding of K6 are linked.
The non-trivial link in K6 may be complicated or simple, and some embeddings will contain more than one non-trivial link. This research project concerns the knotting and linking properties of a typical embedding of a graph. More precisely, after choosing an appropriate random model for graph embeddings, one can analyze the expected values of various numerical characteristics of knots and links inside the graph. For example, in a random linear embedding, the vertices of the graph G are chosen randomly in the unit cube, and edges are taken to be straight line segments between two vertices.
Studying random graph embeddings is analogous to the study of random knots, which has been carried out by a variety of researchers (for example, Arsuaga et al, Panagiotou et al, Pippenger, Diao et al, and many others). The study of random knots is motivated by biology and chemistry, where long, complex molecules such as DNA can become knotted during replication or transcription. Other molecules may have more complex structures that are better modeled as graphs. Certain molecules have been found in specific configurations, but not in others. For example, there are molecules that contain K3,3 subgraphs, and they always appear as Mobius ladders, but not in the configuration containing a trefoil knot. Understanding how random embeddings behave could then shed light on whether this is expected behavior or whether there is biological significance to a particular configuration. With random linear embeddings, it is possible to calculate the likelihood of a random linear embedding of K3,3 appearing in a form isotopic to the Mobius ladder to be approximately 97.38%.

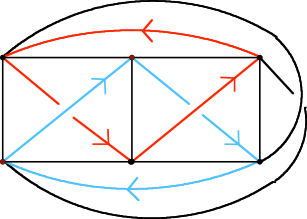
The graph K3,3 embedded as a Mobius ladder.
Potential REU projects will consider questions about further properties of random linear embeddings of complete graphs. Questions that students can investigate include: what is the distribution of knots or links in a random linear embedding of Kn, such as the percentage of cycles or pairs of cycles are unknots or unlinks; and what is the probability that a random linear embedding of Kn contains a link with linking number at least m?
In addition, the project will look at other random embedding models. For knots, it has been experimentally shown that most natural random knot models have the same asymptotic behavior for quantities such as writhe and linking number. Other random graph embeddings have been proposed, and one can also come up with a variety of other invariants for embeddings of graphs that describe their spatial complexity, such as the mean crossing number, which describes the number of times that edges cross in a planar projection, and study their behavior as the number of vertices increases. A comparison of the quantitative behavior of various measurements of topological complexity would be another interesting project to undertake. Questions of interest include: How does the distribution of knots or links in a random embedding of a graph change with respect to the random model? Is there one that is better suited for explicit computation of the distribution of knot and link invariants?